Evaluating a particle size distribution
In general, most particle size distributions that I measured during my career have been best fit by a lognormal distribution. Sometimes one encounters a specimen that is better described by a normal (Gaussian) distribution. It is important to have tools that permit the analyst to scrutinize the results.
In this case our specimen was a dispersion of silver halide particles. These were imaged at -170C in a cooling holder to minimize radiolysis (mass loss of the halide and reduction to Ag0). This is a useful data set, so I included the measurements in my rAnaLab R package.
A representative field is shown below:
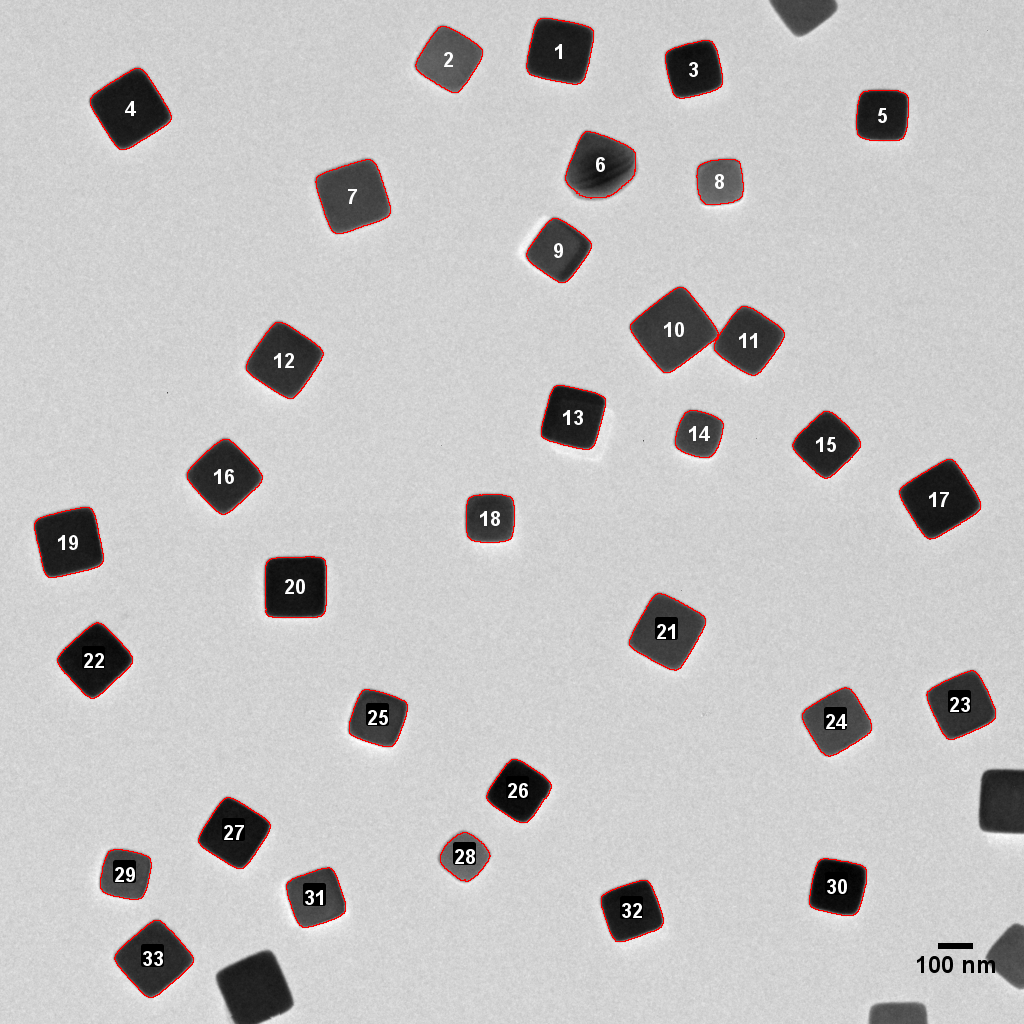
Figure 1: An image of well-dispersed AgX grains imaged at -170C.
We can first compare the parametric statistics from both distributions. The rAnaLab package has a function that makes this convenient.
library(rAnaLab)
data(diam)
ret <- calc.normal.lognormal.stats(diam$ecd.nm)
pander::pander(ret)| nobs | mu | s | geom mean | geom sd |
|---|---|---|---|---|
| 2123 | 131.4 | 17.07 | 130.2 | 1.141 |
Note that the mean value of the two distributions agree within about 1.5 nm. Let’s next look at a panel plot comparing the data to a normal distribution.
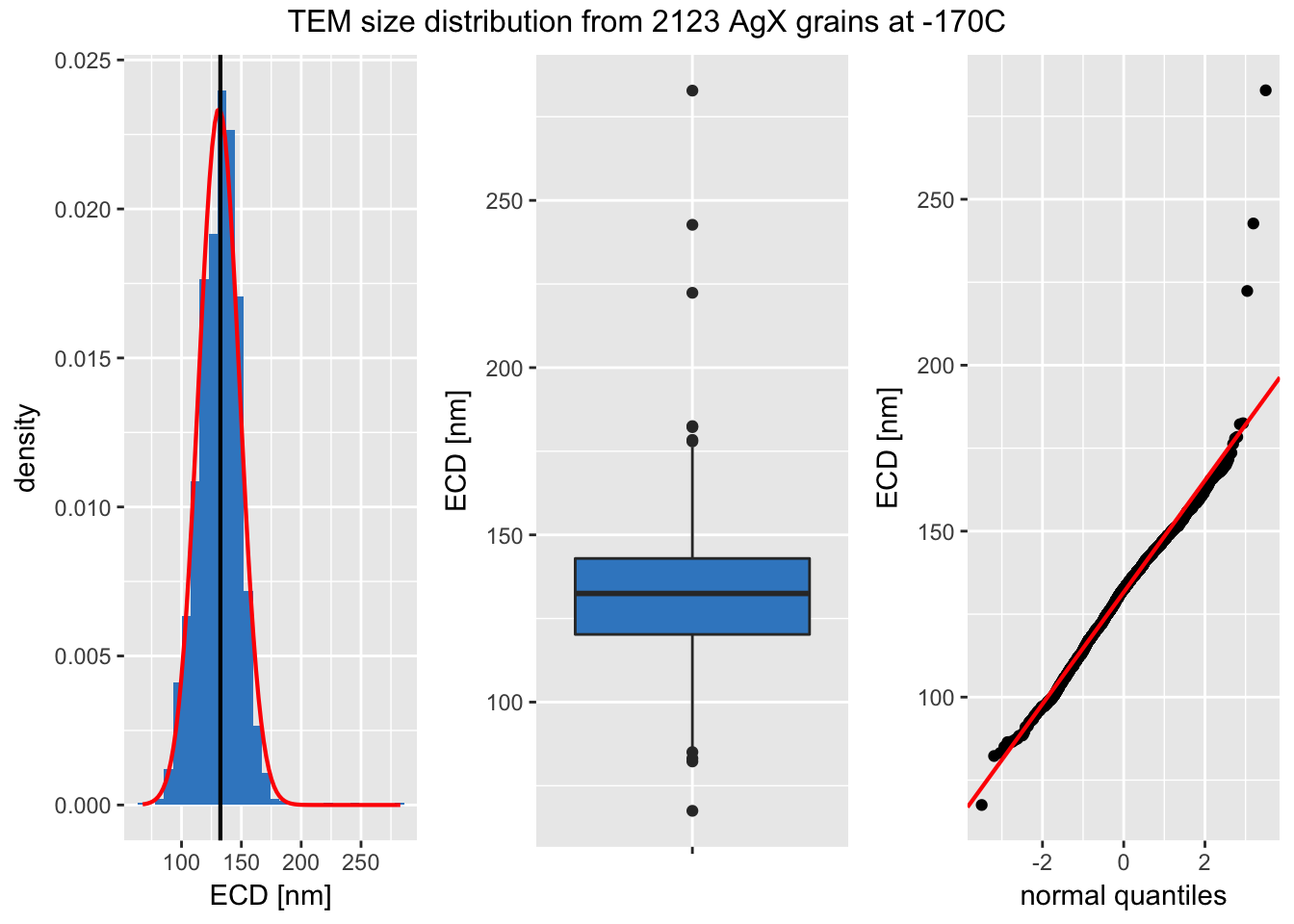
Figure 2: An panel plot from well-dispersed AgX grains imaged at -170C. Left: Histogram and Kernel Density plot. Center: A boxplot highlighting outliers. Right A plot comparing the measured values to a Normal distribution.
I find the panel plot particularly helpful. We only see deviations from the Normal distribution at the extreme tails.
We can now compare to a single mode lognormal distribution. We will first bin the data with a spacing of the 8th root of 2. Again, this has been encapsulated into two function calls. Finally, we will plot the data
l.b <- rb.lognormal.bin(diam[,1], n.root.2=8.0, base.val=10.0)
binned <- make.log.bin.df(l.b)
plt <- ggp.logx.distn(binned,
"AgX Lognormal Grain Diameter",
x.units=-9,
d.max=0.025,
limits=c(50, 500),
breaks=c(50, 100, 500),
minor_breaks=c( 60, 70, 80, 90, 200, 300, 400))
print(plt)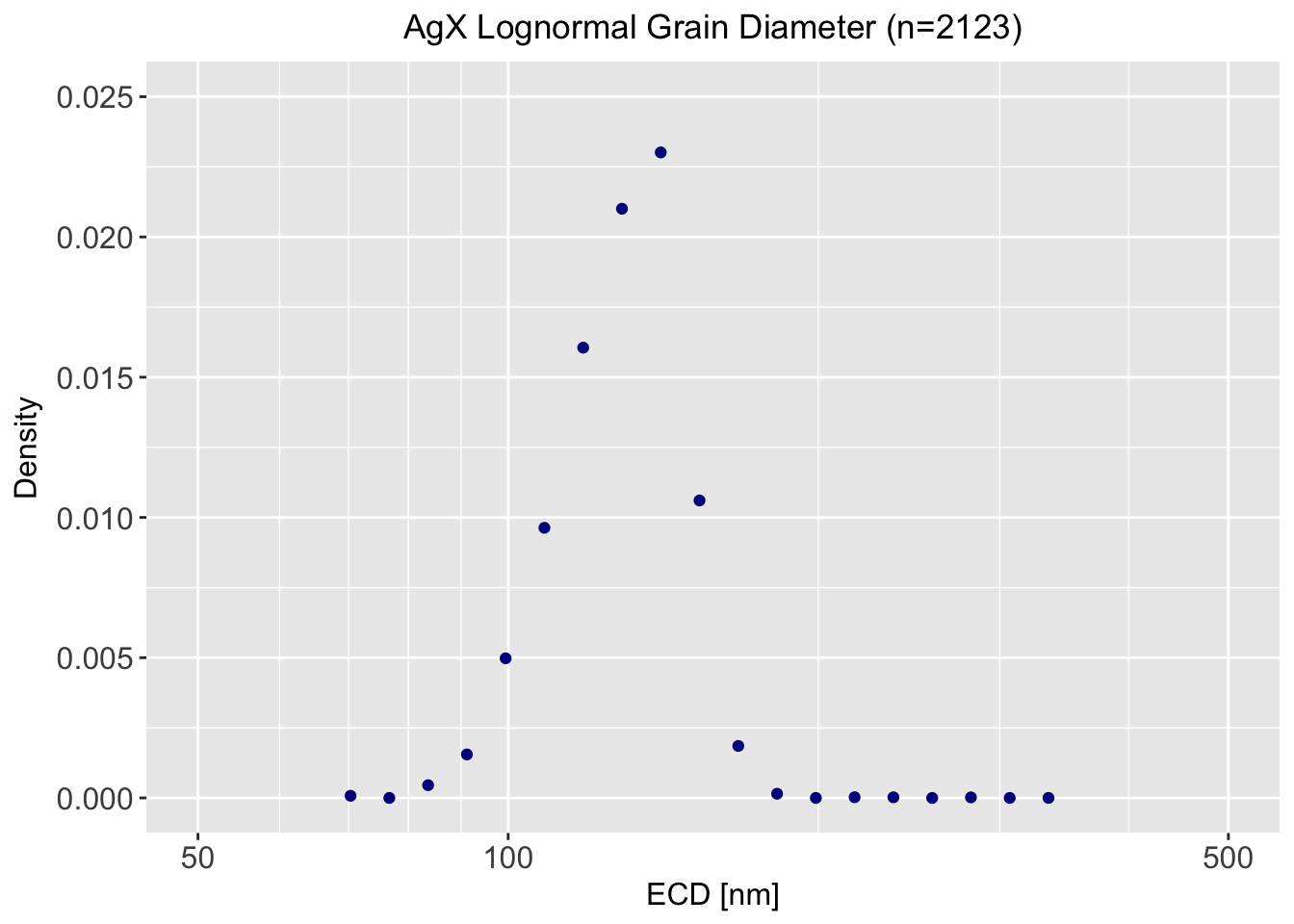
We don’t gain anything in this case by going to a lognormal distribution and it appears slightly skewed. We will follow Occam’s Razor (loosely translates, “It is vain to do with more what can be done with fewer”).